Getting Started with the Latent Labs Platform
Choosing a target
The first step is to choose a protein target to generate binders against. You can do this by:
- Downloading a target from the Protein Data Bank (PDB). Simply choose a PDB ID.
- Uploading a target of your own (CIF is the accepted file format).
- Choosing a past target.
Alternatively, when you log into the platform the first few times, you'll see some example targets to choose from to get up and running quickly.
Configuring your input

-
Select target chain: You will need to specify which chain you'd like your binders to bind to. You can do this by selecting 'Chain A' or 'Chain B' etc. in the sequence view at the bottom of the screen. Alternatively, you can select the chain by clicking on the protein structure itself and selecting a residue.
- Tip✨ We have a 512 residue context limit, which sets a limit on the joint length of the target and binder. You can crop larger targets in the platform by hitting shift + click to select and deselect residues, which also allows to deselect larger stretches of residues in one.
- Tip✨ For large targets, try cropping to a single domain in the target. Domains are compact structural units, which in larger structures can be connected via loops to one another.
- Note It is possible to select multiple chains as target input for the model.
-
Select hotspots Select hotspots to specify where on the chain you would like your binders to bind. You can do this by selecting the specific amino acids in the sequence view at the bottom of the screen. Alternatively, you can select hotspots directly on the chain by clicking on the protein structure itself.
- Tip✨ Hotspots are best selected on the accessible protein surface, and we recommend selecting more than one hotspot. In practice it will not be necessary to select more than a few hotspot residues.
- Tip✨ If your bioassembly contains multiple interacting chains, including interactions with your target of interest, try selecting residues that are bound by another chain already as hotspots. A good way of doing this can be to turn on the 'Atom view' and the 'Sidechains' view, and look at which surface residues on the target make non-covalent bonds with another chain. In the all atom view you can see these bonds visualized via dashed lines.

-
Choose binder length We have successfully lab-validated mini-binders in the length range of 80-120 amino acids, and macrocycles in the length range of 12-18. The platform allows making designs outside of those lab-validated length ranges.

- Choose number of binders Choose 10 or 100 binders as the default options. For more options hit 'advanced settings' where you can input the specific number of binder you would like to generate.
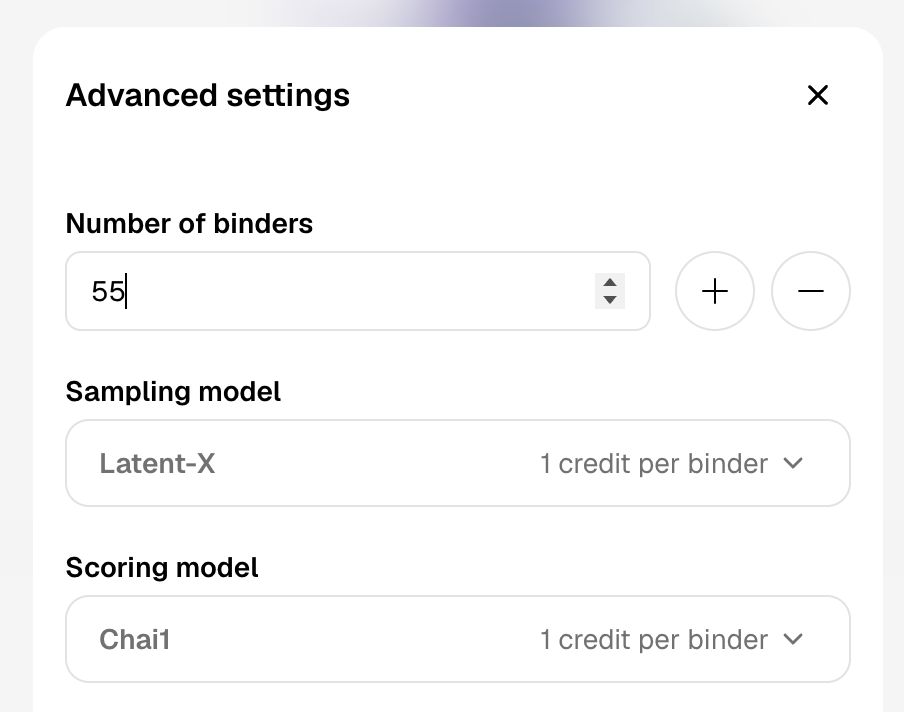
- Hit generateThe time taken to generate and score binders can vary depending on the size of the batch. You can close your browser and come back later - your batch will continue generating and scoring.
Explore, sort and analyse your binders
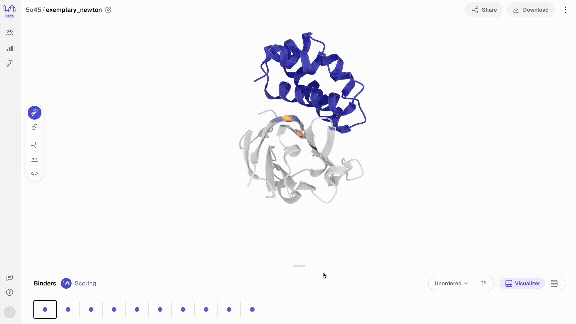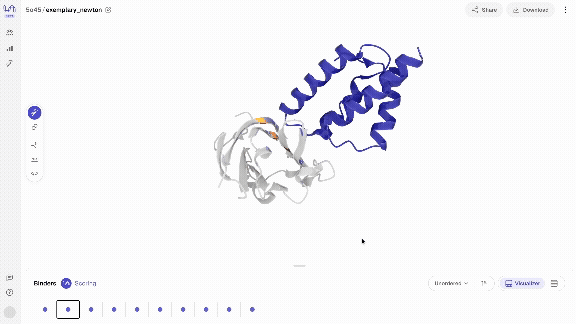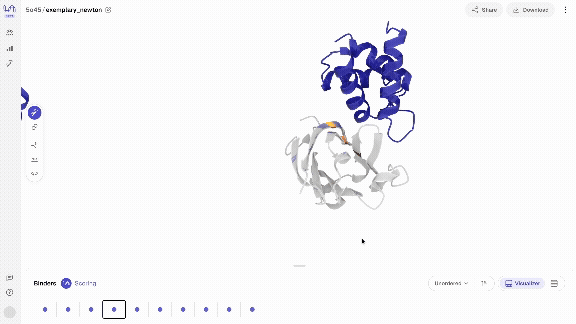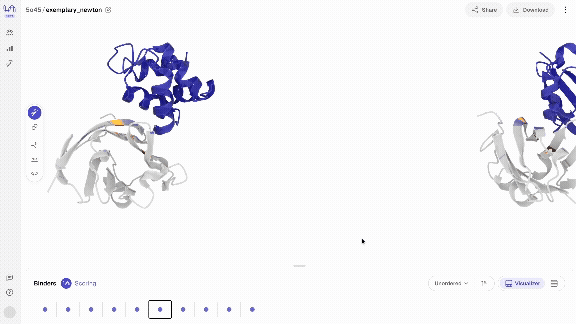
- Scroll through your binders Use your left and right arrow keys to scroll through your binders in the carousel of generated complexes. Your binders will start to appear quickly (usually within seconds) and you can start interacting with them immediately. Scoring your binders may take a bit longer.
- Interact with your binders in different views The model directly generates biochemical bonds such as h-bonds. You can explore different structure views that show this, as well as overlay the generated structures with predicted structures.
- Score & overlay with predicted structure Once scoring has completed, you can overlay the predicted structures by selecting the 'Fold' view. The predicted structures are coloured by pLDDT, a structure prediction confidence metric, where dark blue corresponds to very high, blue to high, yellow to low and orange to very low confidence, resolved per residue.
- Filter and annotate your binders You can sort the binders by different scores, including the 'In-silico filter', a combination of thresholds on structure prediction scores, that we used to select designs for lab validation. You can also annotate and star your binders.
Download and share
Download your binders Hit the download button to receive a .cif file that includes the structure and sequences of all of your binders.
Share with others You can choose to make the project 'publicly' shareable by hitting the share button. This will generate a shareable URL. Individuals who click on the URL will be able to view and download your binders regardless of whether they have platform account. They will not be able to generate their own binders.
Updated 3 months ago
Check out our FAQs for further support
